
The Poirier Lab

 |
The Poirier Lab |
 |
| Main Page | Research | People | News | Publications | Teaching | Positions Available | Contact Information | Share Drive |
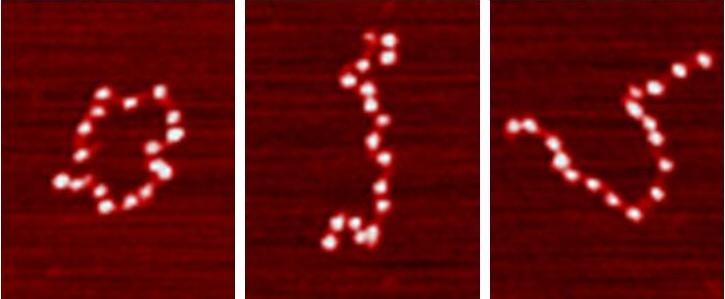
ResearchModern biological physics and molecular biology aims to reveal the molecular mechanisms of biological functions such as the expression of a gene and the repair of a damaged genome. Our genomes are organized into chromosomes, which contain long chromatin fibers that are a complex assortment of DNA and protein. The fundamental unit of chromatin is the nucleosome, which contains ~147 base pairs of DNA wrapped around an octamer of H2A, H2B, H3, and H4 histone proteins. This results in our genomic DNA being organized into ~20 million nucleosome spools. Nucleosome and chromatin structure and dynamics play central roles in regulating all forms of DNA processing, including RNA transcription and DNA repair. We are quantitatively investigating the mechanisms by which our genome is expressed and repaired. Our approach is to apply a cross-disciplinary techniques that combines biochemical, biophysical and single molecule techniques, which includes: restriction enzyme studies, cyclization experiments, nucleosome mapping, steady state Fluorescence Resonance Energy Transfer measurements (FRET), stopped flow FRET, fluorescence correlation spectroscopy, single molecule force and twist measurements with magnetic tweezers, Atomic Force Microscopy (AFM) measurements and single molecule FRET measurements. Because of our interdisciplinary
nature, collaborations are essential for our success. We have developed
close collaborations with the Ottesen
Lab
and the Bundschuh
Lab. Nucleosome Structure and FunctionHistones function to organize and compact our genomic DNA into nucleosomes and chromatin while histone modifications function to facilitate and regulate biological process that involve DNA-protein interactions, such as RNA transcription and DNA repair. The nucleosome contains 2 of each core histone protein: H2A, H2B, H3 and H4, and 147 base pairs of DNA This structure is densely repeated along our genomic DNA so that our chromosomes contain about 20 million nucleosomes, and creates long chromatin fibers. There are over 100 known post-translational histone modifications; however the molecular functions of most of these modifications are not well understood.Histone modifications are thought to function in one of two ways: (i) to create a protein binding site and (ii) to directly modify the structure and/or dynamics of chromatin. A majority of modifications are located on histone tails, which are unstructured regions of the nucleosome and emanate from the nucleosome core making the tails accessible for protein binding. However, recent mass spectrometry studies have revealed that there are over 30 modifications in the structured region of the nucleosome and 15 of them are located within the DNA-histone interface of the nucleosome. My lab is focused on four histone H3 modifications that are positioned within structured regions of the nucleosome. We are experimentally
investigating models of how histone modifications function while
buried within the DNA histone interface of the nucleosomes. We
collaborate with Prof.
Jennifer Ottesen (OSU), who constructs homogeneously modified
histone H3 proteins using Expressed Protein Ligation, which is a
powerful technique for the introduction of chemical modifications into
one region of a protein. We are investigating molecular models of PMT
function with a multidisciplinary approach that includes the
restriction enzyme kinetics method, nucleosome mapping, histone binding
affinity measurements, steady state Fluorescence Resonance Energy
Transfer measurements (FRET), stopped flow FRET, fluorescence
correlation spectroscopy, single molecule force and twist measurements
with magnetic tweezers, and single molecule FRET measurements.
DNA and Chromatin Mechanics
|
| Main Page | Research | People | News | Publications | Teaching | Positions Available | Contact Information | Share |
This page last modified on July 16, 2008.